Looking for what that is not there
“If I had eight hours to chop down a tree, I’d spend six sharpening my axe.” - Abraham Lincoln via Kline (2016). This adage is appropriate to set the tone for this post, as well as applicable to most things in general, including working with data.
My professors taught me that real data never works, and my experience attested to their statement countless times as I iterated over the data work procedure of importing, cleaning, model building, model tuning, and communicating results. One thing about it that I used to find frustrating is the data I got if oftentimes incomplete (or partly missing).
Missing data are values that should have been recorded but were not. The best way to treat missing data is not to have them, but unfortunately, real data is oftentimes
uglyunorganized. Missing data could potentially caused by nonresponse in surveys, or technical issues with data-collecting equipment.My previous posts were about visualizing data that we have, but this time, we will be visualizing things that we ‘do not’ have (aka missing data), as well as discussing about ways we can deal with them via complete case analysis or imputation.

Import and read the data set
- As usual, we will begin by importing essential libraries and load in the data set to preprocess it.
Show code
library(foreign) #To read SPSS data
library(tidyverse) #datawork toolbox
library(dlookr) #for missing data diagnosis
library(visdat) #for overall missingness visualization
library(naniar) #for missingness visualization
library(VIM) #for donor-based imputation
library(simputation) #for model-based imputation
library(mice) #for multiple imputation
Show code
#Import the data set
PISA_CAN <-read.spss("PISA2018CAN.sav",to.data.frame = TRUE, use.value.labels = FALSE)
#Subset and rename the variables
PISA_Subsetted <- PISA_CAN %>%
select(REPEAT, FEMALE = ST004D01T, ESCS, DAYSKIP = ST062Q01TA,
CLASSSKIP = ST062Q02TA, LATE = ST062Q03TA,
BEINGBULLIED, DISCLIMA, ADAPTIVITY)
#Recode variables into factor
PISA_Subsetted$DAYSKIP <-as.factor(PISA_Subsetted$DAYSKIP)
PISA_Subsetted$CLASSSKIP <-as.factor(PISA_Subsetted$CLASSSKIP)
PISA_Subsetted$LATE <-as.factor(PISA_Subsetted$LATE)
PISA_Subsetted$FEMALE <-as.factor(PISA_Subsetted$FEMALE)
PISA_Subsetted$REPEAT <-as.factor(PISA_Subsetted$REPEAT)
# Renaming factor levels with dplyr
PISA_Subsetted$FEMALE <- recode_factor(PISA_Subsetted$FEMALE,
"1" = "1", "2" = "0")
glimpse(PISA_Subsetted)
Rows: 22,653
Columns: 9
$ REPEAT <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,~
$ FEMALE <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,~
$ ESCS <dbl> -0.7302, 0.3078, 0.5059, 1.1147, 1.3626, -0.857~
$ DAYSKIP <fct> 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 3, 1, 1, 2, 1, 2,~
$ CLASSSKIP <fct> 1, 1, 1, 1, 1, 2, 2, 1, 1, 1, 3, 1, 1, 2, 2, 2,~
$ LATE <fct> 2, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 2, 1, 3,~
$ BEINGBULLIED <dbl> 1.2618, 1.7669, 0.1462, -0.7823, 0.2907, 0.7703~
$ DISCLIMA <dbl> -0.4186, -1.4179, 0.6019, -0.4995, -0.1045, 1.0~
$ ADAPTIVITY <dbl> -0.4708, 0.6350, -0.5786, -0.5786, -0.0763, 0.5~The data set in this post was a Canadian student data subsetted from the Programme for Internal Student Assessment (PISA), which is an international assessment that measures 15-year-old students’ reading, mathematics, and science literacy every three years.
From the
glimpsecall above, our dataset has 9 variables and 22,653 data points.
Check for missing data
First, we will use the
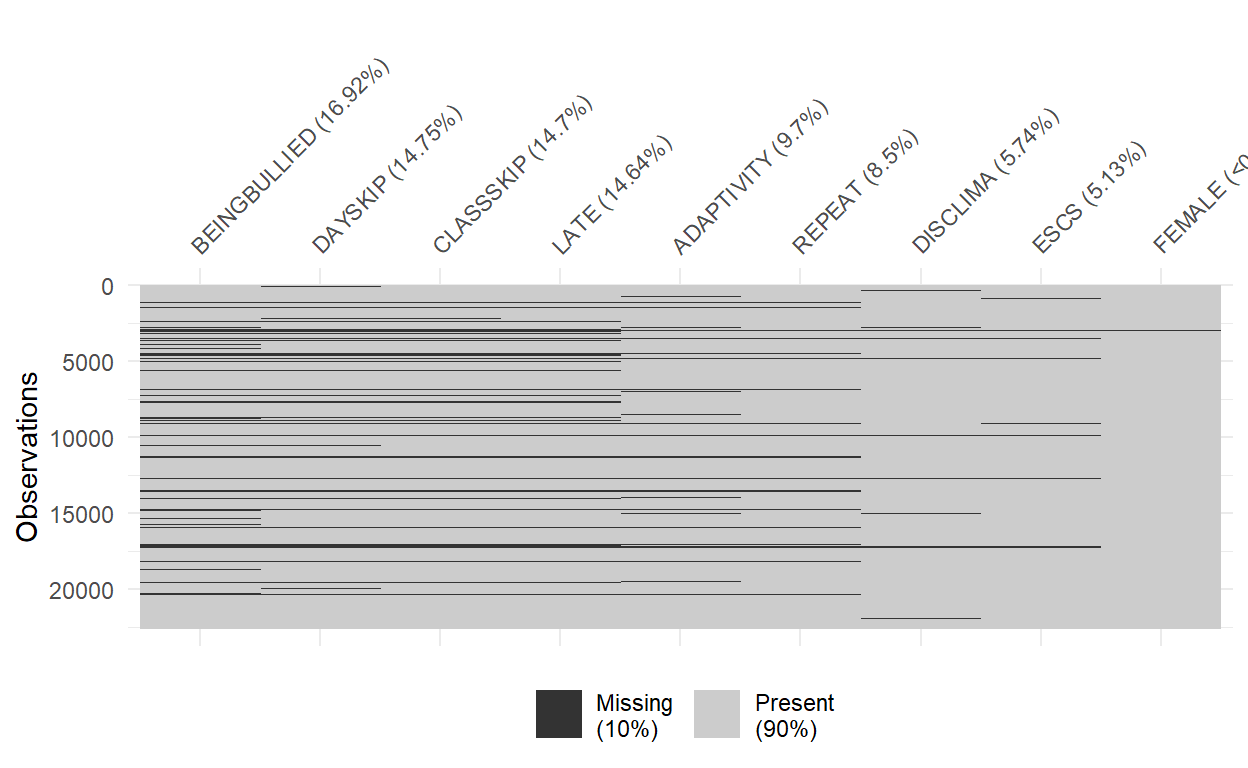
dlookrpackage to diagnose missingness of the data set, as well as plot missing data map withvis_miss.The plot provides a specific visualization of the amount of missing data, showing in black the location of missing values, and also providing information on the overall percentage of missing values overall (in the legend), and in each variable.
Show code
dlookr::diagnose(PISA_Subsetted)
# A tibble: 9 x 6
variables types missing_count missing_percent unique_count
<chr> <chr> <int> <dbl> <int>
1 REPEAT factor 1926 8.50 3
2 FEMALE factor 2 0.00883 3
3 ESCS numeric 1163 5.13 15366
4 DAYSKIP factor 3341 14.7 5
5 CLASSSKIP factor 3330 14.7 5
6 LATE factor 3316 14.6 5
7 BEINGBULLIED numeric 3833 16.9 63
8 DISCLIMA numeric 1300 5.74 934
9 ADAPTIVITY numeric 2197 9.70 65
# ... with 1 more variable: unique_rate <dbl>Show code
visdat::vis_miss(PISA_Subsetted, sort_miss = TRUE)
- If we are curious about the proportion of missing data by groups, we can also group the dataset by our categorical variable of interest, say, gender, before examining the missingness ratio.
Show code
PISA_Subsetted %>% group_by (FEMALE) %>%
miss_var_summary()
# A tibble: 24 x 4
# Groups: FEMALE [3]
FEMALE variable n_miss pct_miss
<fct> <chr> <int> <dbl>
1 1 BEINGBULLIED 1651 14.6
2 1 DAYSKIP 1451 12.8
3 1 CLASSSKIP 1445 12.8
4 1 LATE 1430 12.6
5 1 ADAPTIVITY 907 8.02
6 1 REPEAT 812 7.18
7 1 DISCLIMA 563 4.98
8 1 ESCS 527 4.66
9 0 BEINGBULLIED 2180 19.2
10 0 DAYSKIP 1888 16.6
# ... with 14 more rowsTypes of Missing Data
Yes, we know now that our data is missing, but not all missing data are created (or not created, pun wholeheartedly intended) equal. There are three types of missing data, MCAR, MAR, and MNAR.
Missing Completely at Random (MCAR)
Locations of missing values in the dataset are purely random. they do not depend on any other data.
For example, if a doctor forgets to record the age of every tenth patient entering an ICU, the presence of missing value would not depend on the characteristic of the patients.
Missing at Random (MAR)
Locations of missing values in the dataset depend on some other, observed data.
Data are considered as MAR if the probability of missingness is unrelated to the actual value on that variable after controlling for the other variables in the dataset
In survey data, high-income respondents are less likely to inform the researcher about the number of properties owned.
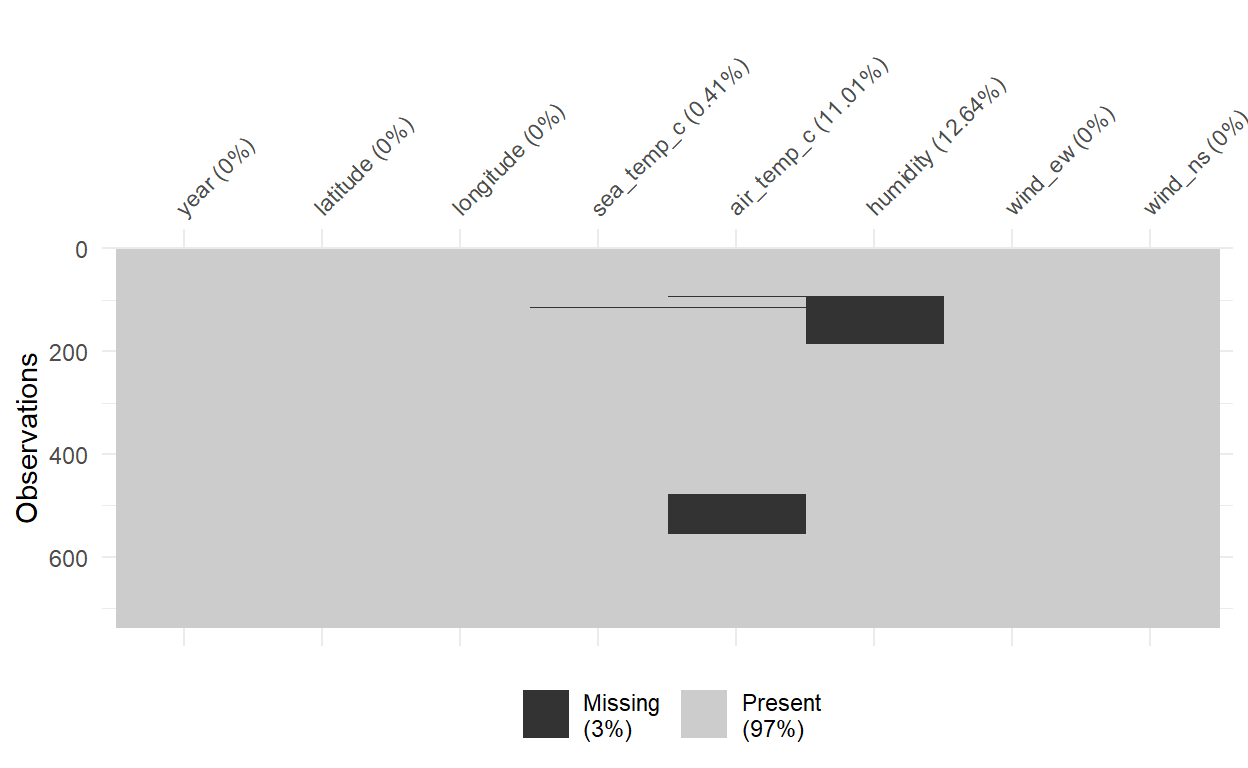
Below is an example of MAR missingness. See that
sea_tempandair_tempare missing at a certain part of the year. Maybe the measuring tools broke down or something before they got them fixed.
Missing Not at Random (MNAR)
If it is not MCAR or MAR, it is probably MNAR. This is the most tricky type of missingness to handle.
The missing values depend on both characteristics of the data and also on missing values. In this case, determining the mechanism of the generation of missing value is difficult.
Missing values for a variable like blood pressure may partially depend on the values of blood pressure as patients who have low blood pressure are less likely to get their blood pressure checked at frequently.
Visualize Missing data
- Okay, now we know what missing data is, and what are types of missing data, here are some ways we can visualize them so that we know their patterns and what they are up to.
Missing pattern w/Upset plot
- An upset plot from the
UpSetRpackage can be used to visualize the patterns of missingness, or rather the combinations of missingness across cases and variables.
Show code
gg_miss_upset(PISA_Subsetted, nsets = 9, nintersects = 15)
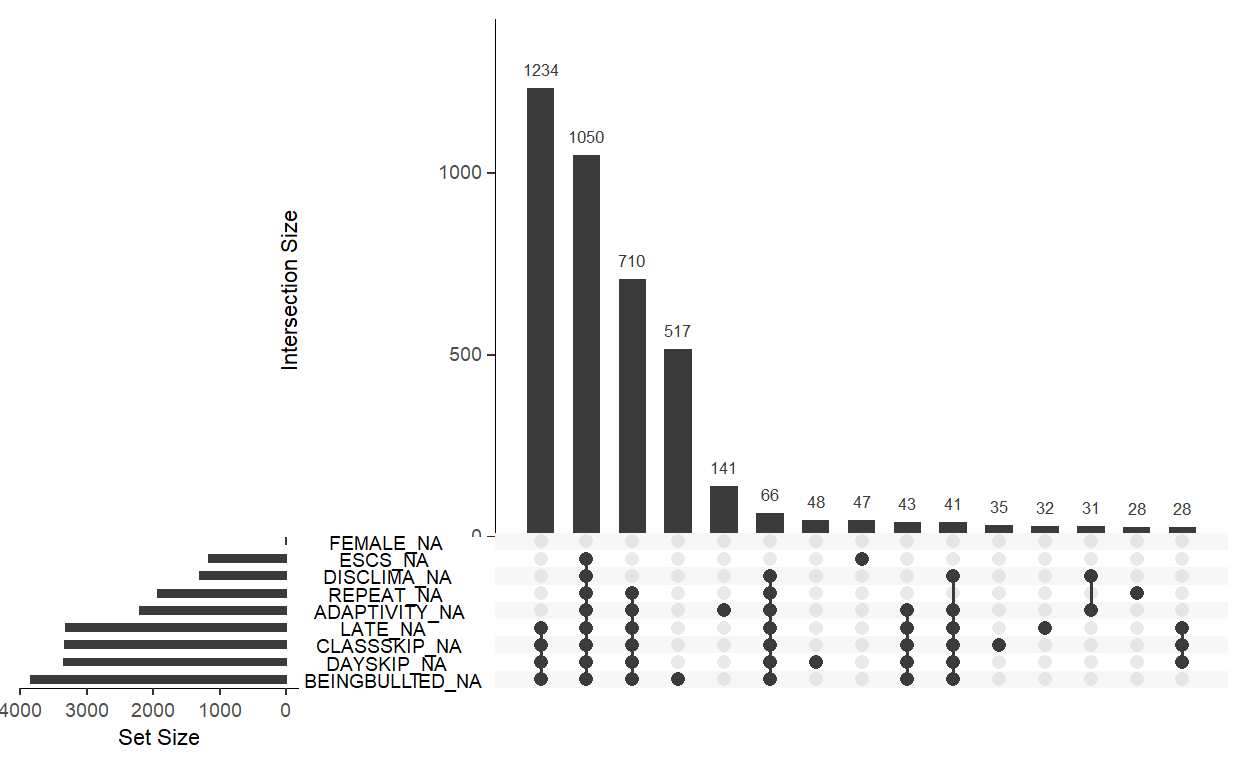
The small bar plot to the left indicated the amount of missingness in variables. Consistent with the missingness diagnosis, the variable
BEINGBULLIEDhas the most missing data, following byDAYSKIPandCLASSKIP.The dot plot to the right showed combinations of variable that are missing in the data set. For example, there are 1,234 cases that have missing data in the variable
LATE,CLASSKIP,DAYSKIP, andBEINGBULLIED.The parameter
nsetslooks at 9 sets of variables, while the parameternintersectslooks at 15 variable combinations.
General visual summaries of missing data
- This section demonstrates numerous ways to visualize missing data to determine their patterns.
Missingness in variables with gg_miss_var
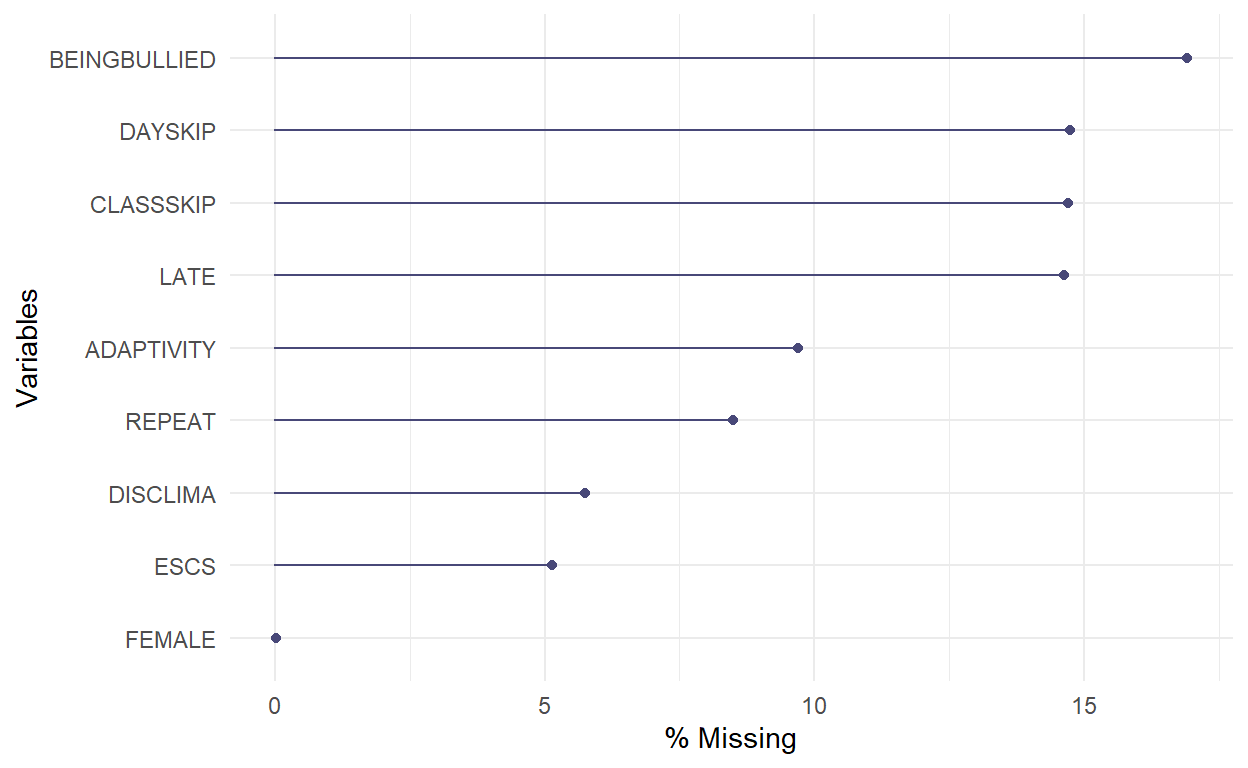
- This plot shows the number of missing values in each variable in a dataset.
Show code
PISA_Subsetted %>% miss_var_table()
# A tibble: 9 x 3
n_miss_in_var n_vars pct_vars
<int> <int> <dbl>
1 2 1 11.1
2 1163 1 11.1
3 1300 1 11.1
4 1926 1 11.1
5 2197 1 11.1
6 3316 1 11.1
7 3330 1 11.1
8 3341 1 11.1
9 3833 1 11.1Show code
gg_miss_var(PISA_Subsetted, show_pct = TRUE)
Missingness in cases with gg_miss_case
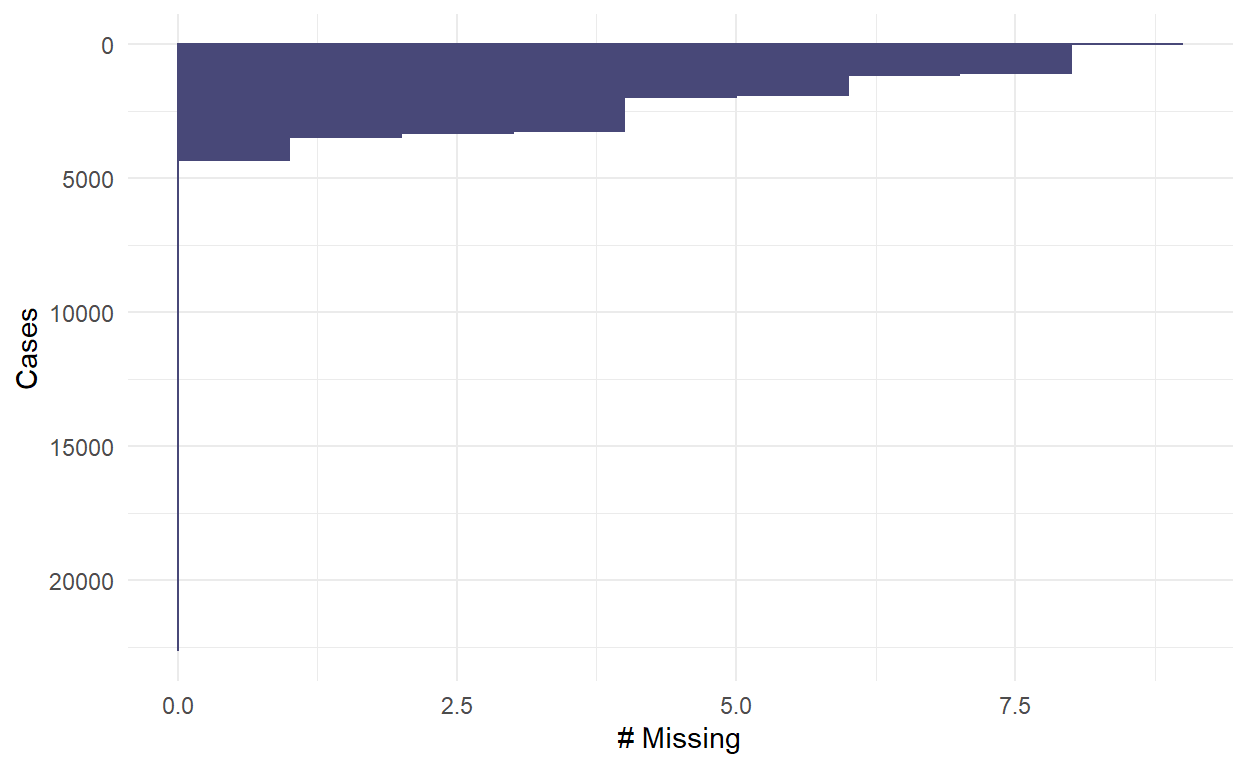
- This plot shows the number of missing values in each case. For example, the table showed that there are 2 cases with 9 missing variables (i.e., no data in all variables), and there are 1050 cases with 8 missing variables.
Show code
PISA_Subsetted %>% miss_case_table()
# A tibble: 10 x 3
n_miss_in_case n_cases pct_cases
<int> <int> <dbl>
1 0 18327 80.9
2 1 870 3.84
3 2 140 0.618
4 3 81 0.358
5 4 1254 5.54
6 5 83 0.366
7 6 756 3.34
8 7 90 0.397
9 8 1050 4.64
10 9 2 0.00883Show code
gg_miss_case(PISA_Subsetted)
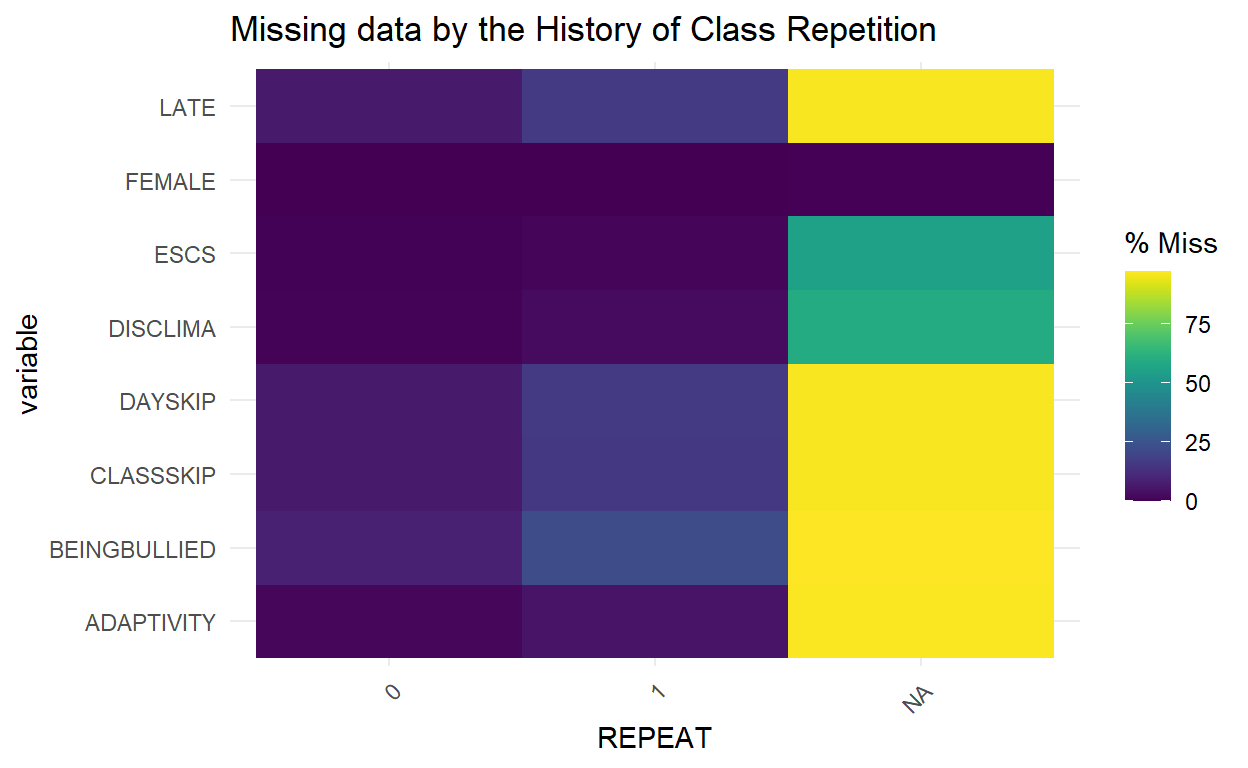
Missingness across factors with gg_miss_fct
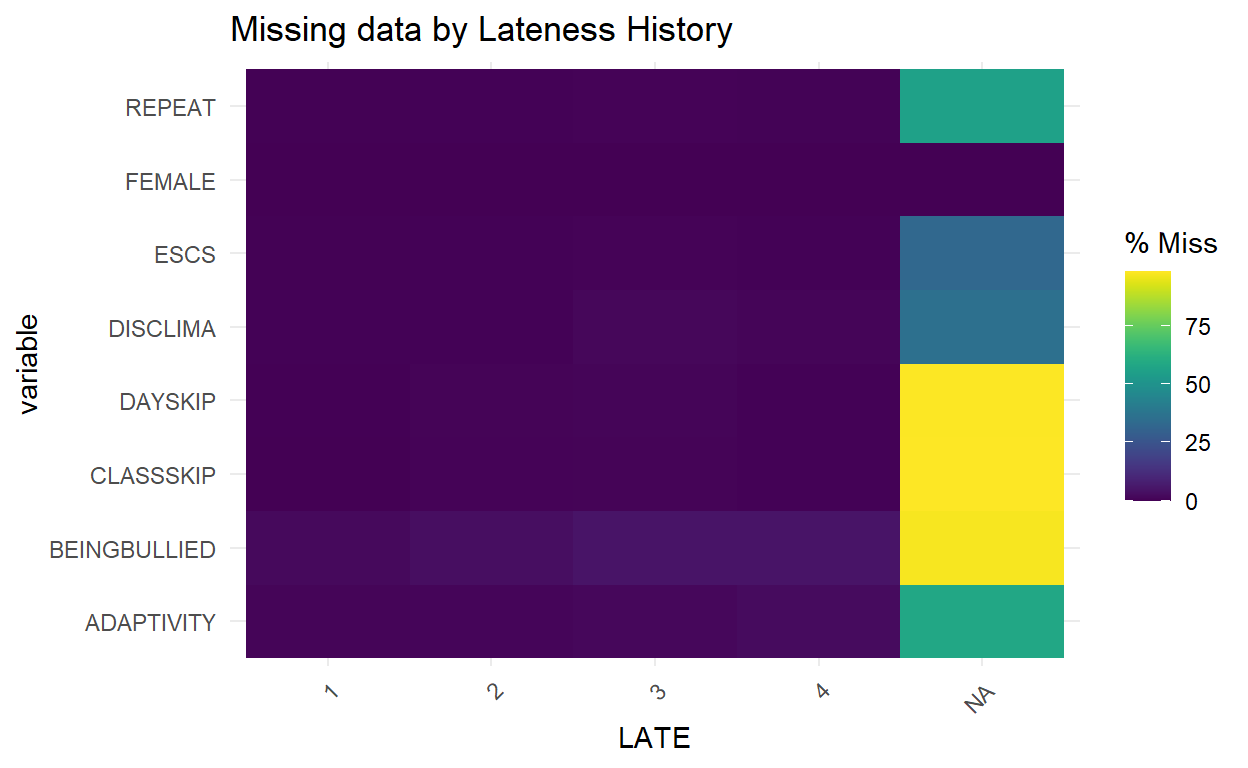
- This plot shows the number of missingness in each column, broken down by a categorical variable from the dataset.
Show code
gg_miss_fct(x = PISA_Subsetted, fct = REPEAT) +
labs(title = "Missing data by the History of Class Repetition")
Show code
gg_miss_fct(x = PISA_Subsetted, fct = LATE) +
labs(title = "Missing data by Lateness History")
- The heatmap above showed the proportion of missing data we have in each response of the selected categorical variable; for example, the history of class repetition (
REPEAT), with0as no,1as yes, andNAas missing.
Missingness along a repeating span with gg_miss_span
- This plot showed the number of missings in a given span, or breaksize, for a single selected variable.
Show code
gg_miss_span(PISA_Subsetted, REPEAT, span_every = 2000) +
theme_dark()
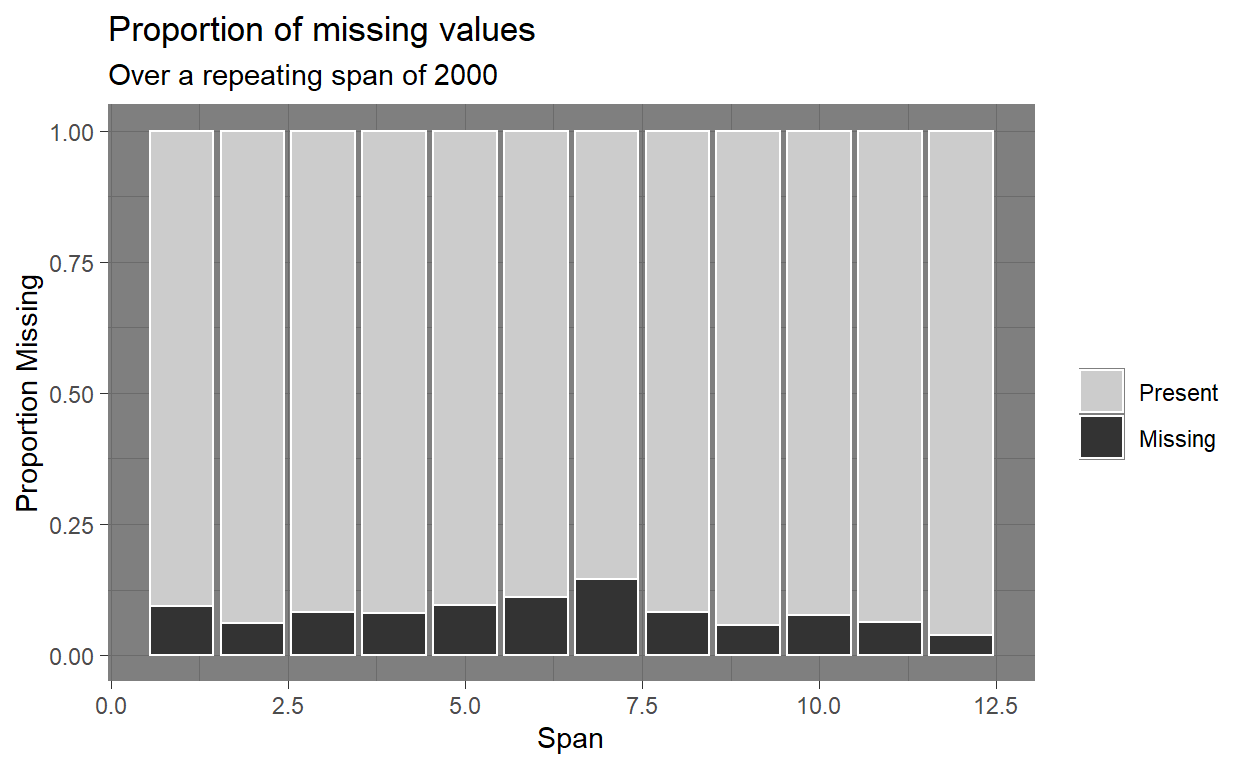
- The plot went over the data and showed us how many missing data we have every 2000 data points that it went through.
Cumulative missing
- This plot showed the cumulative amount of missing value over the data set. A sharp increase in cumulative missing value could indicate missing patterns to be discovered.
Show code
PISA_Subsetted %>% gg_miss_case_cumsum(breaks = 2000) + theme_bw()
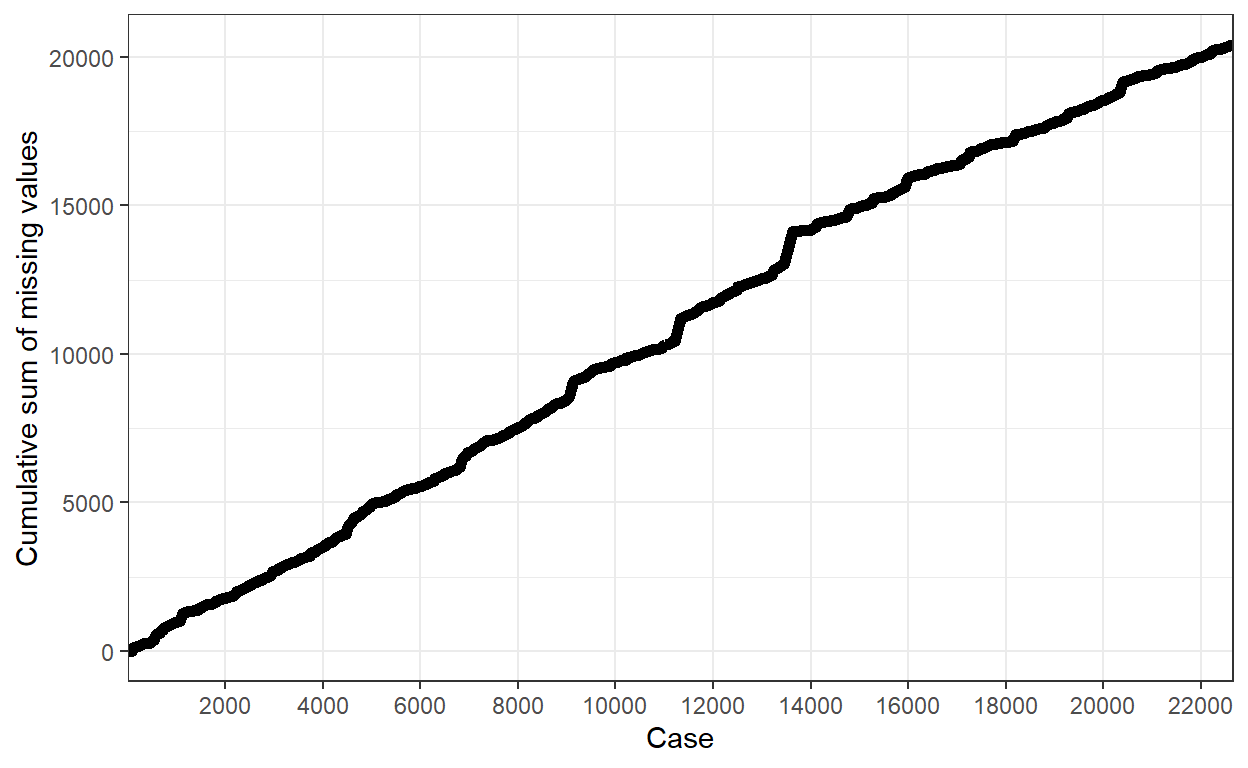
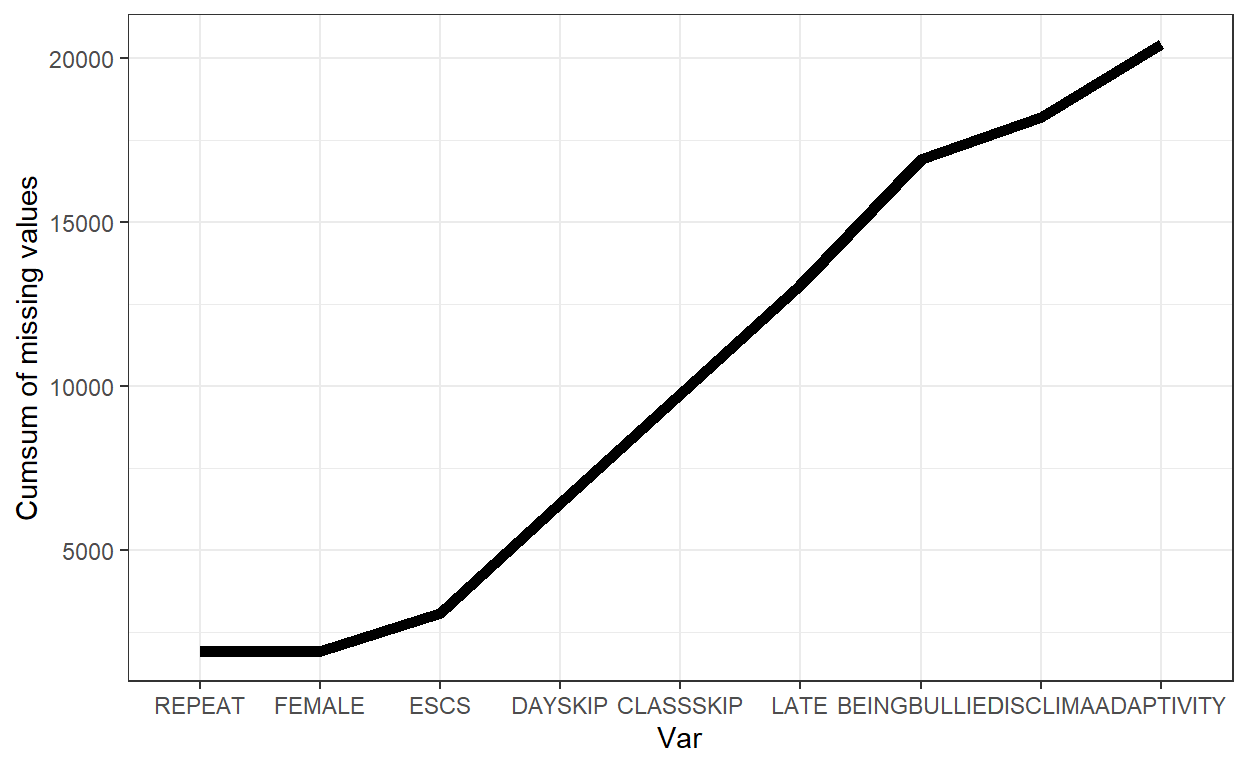
- This plot showed the cumulative amount of missing value over the variable. We could examine the relative proportion of missing values across variables via this plot.
Show code
PISA_Subsetted %>% gg_miss_var_cumsum() + theme_bw()
What should we do with the missing data
Now that we know we have missing data, there are numerous ways we can deal with it such as disregarding them with complete case analysis, or making educated guesses with imputation.
Anyway, dealing with missing data helps minimizing bias in the data, maximizing the use of available information (We don’t want to throw away any of our hard-earned data), and increasing the chance of getting a good reliability estimates such as standard errors, confidence intervals, and p-values.
Complete Case Analysis
Listwise deletion is the method of deleting all cases with missing value, so that we get a clean and complete data set as a result, at the expense of losing a chunk of data in the process.
- Listwise deletion is often a default way to handle missing data (e.g., SPSS).
- This often results in losing 20% to 50% of the data.
- Listwise deletion is often a default way to handle missing data (e.g., SPSS).
Show code
PISA_Listwise <- PISA_Subsetted[complete.cases(PISA_Subsetted), ]
glimpse(PISA_Listwise)
Rows: 18,327
Columns: 9
$ REPEAT <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,~
$ FEMALE <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,~
$ ESCS <dbl> -0.7302, 0.3078, 0.5059, 1.1147, 1.3626, -0.857~
$ DAYSKIP <fct> 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 3, 1, 1, 2, 1, 2,~
$ CLASSSKIP <fct> 1, 1, 1, 1, 1, 2, 2, 1, 1, 1, 3, 1, 1, 2, 2, 2,~
$ LATE <fct> 2, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 2, 1, 3,~
$ BEINGBULLIED <dbl> 1.2618, 1.7669, 0.1462, -0.7823, 0.2907, 0.7703~
$ DISCLIMA <dbl> -0.4186, -1.4179, 0.6019, -0.4995, -0.1045, 1.0~
$ ADAPTIVITY <dbl> -0.4708, 0.6350, -0.5786, -0.5786, -0.0763, 0.5~Notice that the size of our dataset got reduced to 18,327 cases. This happaned from deleting all cases with missing value.
Pairwise deletion is the method that deletes cases only if they have missing data on variables involved in a particular computation, so we can still retain the data for other analyses that do not involve variables that are missing. However, the effective sample size can vary from one analysis to another.
As a demonstration, we will calculate a covariance matrix using pairwise complete observation method.
Show code
BEINGBULLIED DISCLIMA
BEINGBULLIED 1.1315670 -0.1982891
DISCLIMA -0.1982891 1.1373620HOWEVER, the bias caused by using listwise/pairwise deletion has been shown in simulations to grossly exaggerate or underestimate some effects.
Despite giving valid estimates when data are MCAR, the statistical power will be severely reduced when there is a lot of missingness. If the missingness is MAR or MNAR, removing them introduces bias to models built on these data.
Missing Data Imputation
- Other than disregarding them, we can replace the missing value with our best guess with imputation. There are three approaches we can use, donor-based imputation, model-based imputation, and multiple imputation.
Donor-based imputation
- Donor-based imputation replaces missing values based on other complete observations.
Mean Imputation
Mean imputation replaces all missing values with the mean of that variable.
First, we will create a binary indicator for whether each value was originally missing.
Show code
DISCLIMA_imp ADAPTIVITY_imp
1 FALSE FALSE
2 FALSE FALSE
3 FALSE FALSE
4 FALSE FALSE
5 FALSE FALSE
6 FALSE FALSE- Replace missing values in DISCLIMA and ADAPTIVITY variables with their respective means.
Show code
DISCLIMA ADAPTIVITY DISCLIMA_imp ADAPTIVITY_imp
1 -0.4186 -0.4708 FALSE FALSE
2 -1.4179 0.6350 FALSE FALSE
3 0.6019 -0.5786 FALSE FALSE
4 -0.4995 -0.5786 FALSE FALSE
5 -0.1045 -0.0763 FALSE FALSE
6 1.0832 0.5464 FALSE FALSE- We can try plotting the data on a margin plot to see the result of our mean imputation.
Show code
PISA_meanimp %>% select(DISCLIMA, ADAPTIVITY, DISCLIMA_imp, ADAPTIVITY_imp) %>% marginplot(delimiter="imp")
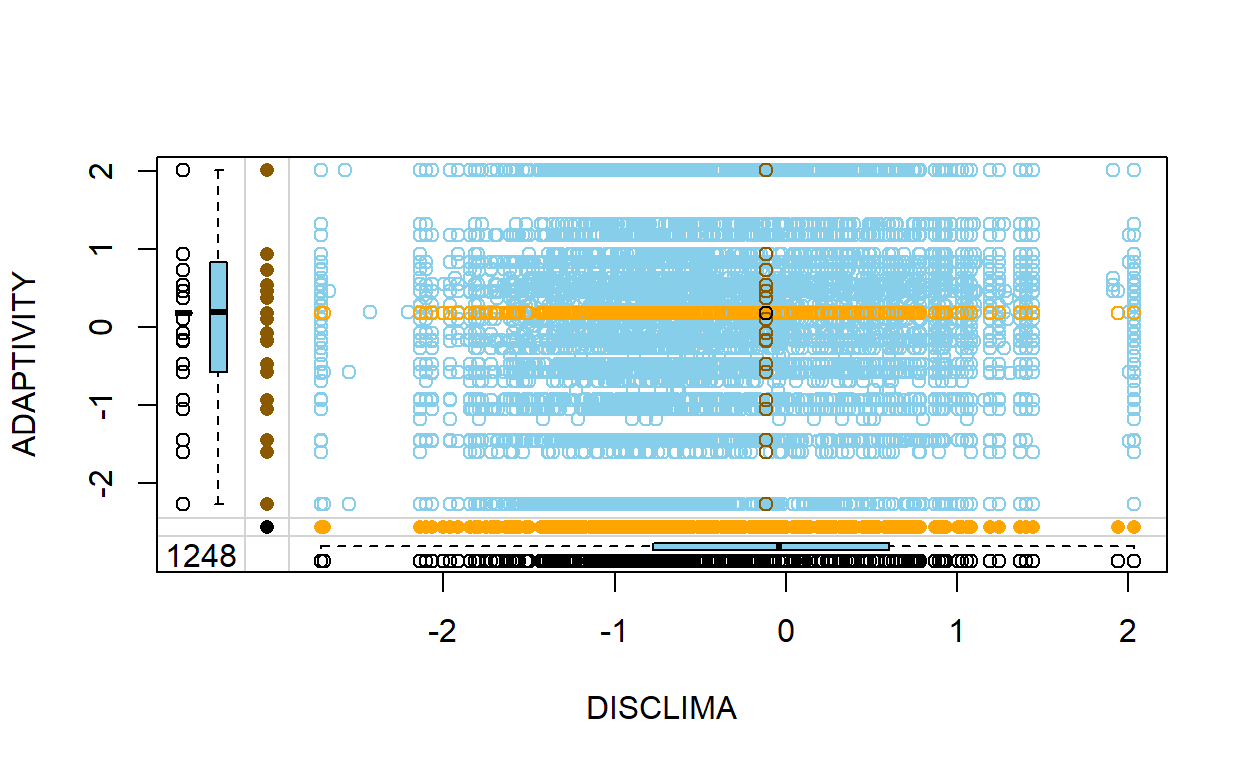
You can see that all missing values were replaced by the mean of that variable. Yes, we got the data back, but what did it cost?
Mean imputation destroys relationship between variables.
Models predicting one using the other will be fooled by the outlying imputed values and will produce biased results.
Mean imputation
crushestakes away variance in the data, which could potentially underestimate all standard errors. This prevents reliable hypothesis testing and calculating confidence interval.This method is not generally recommended, but to each their own. Use it at your own discretion. With the right justification from the literature, mean imputation can be a viable method in your analysis as well.
K-Nearest Neighbor(kNN) Imputation
For kNN imputation, we identify ‘k’ samples in the dataset that are similar or close in the space. Then we use these ‘k’ samples to estimate the value of the missing data points.
Basically, it is like you have a data point with missing values asks its neighbors what value do they have on the variable that it is missing. That data point then replace its missing values with the value of its nearest neighbor.
Show code
DISCLIMA ADAPTIVITY DISCLIMA_imp ADAPTIVITY_imp
1 -0.4186 -0.4708 FALSE FALSE
2 -1.4179 0.6350 FALSE FALSE
3 0.6019 -0.5786 FALSE FALSE
4 -0.4995 -0.5786 FALSE FALSE
5 -0.1045 -0.0763 FALSE FALSE
6 1.0832 0.5464 FALSE FALSE- Note that there are two more columns added, DISCLIMA_imp and ADAPTIVITY_imp. The two added columns tell us if our variables of interest were imputed with the kNN method or not, with
TRUEindicates that the value was imputed andFALSEindicates otherwise.
Model-based imputation
For model-based imputation, missing values are predicted with a statistical or machine learning model.
The model that we used depends on the type of the missing variable:
- Continuous variables - linear regression
- Binary variables - logistic regression
- Categorical variables - multinomial logistic regression
- Count variables - Poisson regression
Linear Regression Imputation
Show code
REPEAT FEMALE ESCS DAYSKIP CLASSSKIP
1926 2 1163 3341 3330
LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3316 3833 1278 2056 - Notice that we have managed to impute some cases of
DISCLIMAandADAPTIVITYbased on the availability of other variables in the same case. However, if there is no other variable on that case (i.e., complete missing), the model won’t be able to predict the target value as there is no predictor available.
Logistic Regression Imputation
- Logistic regression imputation is similar to linear regression imputation, with a difference in the nature of missing value (Continuous vs Binary).
[1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[12] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSEShow code
PISA_logregimp <- PISA_Subsetted
logreg_model <- glm(REPEAT ~ DAYSKIP + BEINGBULLIED + ESCS,
data = PISA_Subsetted, family = binomial)
preds <- predict(logreg_model, type = "response")
preds <- ifelse(preds >= 0.5, 1, 0)
PISA_logregimp[missing_REPEAT, "REPEAT"] <- preds[missing_REPEAT]
table(preds[missing_REPEAT])
0 1
1669 1 Show code
table(PISA_Subsetted$REPEAT)
0 1
19575 1152 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP
256 2 1163 3341 3330
LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3316 3833 1300 2197 Multiple Imputation by Chained Equation
Multiple Imputation by Chained Equation (MICE) - also known as sequential regression multiple imputation - is an emerging method in dealing with missing values by implementing the imputation multiple times as opposed to the single imputation methods mentioned above (Azur et al., 2011).
With the right model, MICE was found to be effective in reducing bias, especially in a large data set with MCAR and MAR. The method basically imputed the missing value with a statistical model (say, linear regression) multiple times for different imputed values before pooling the results together for the final most likely value that the algorithm can come up.
The package
micehas several statistics and machine learning models we can use such as Predictive mean matching (pmm), Classification and Regression Tree (cart), and Random Forest Imputation (rf). Keep in mind that it is a best practice to justify our selected model in missing data imputation to make the analysis as less ‘black box’ as possible for explainability.For this post, I will use the predictive mean matchmaking method that calculates the predicted value from a randomly drawn set of candidate donors that have the value closest to the missing entry. The assumption is the distribution of the missing cell is the same as the observed data of the candidate donors.
The rationale is that PMM produces little biased estimates when missing data is below 50% and not systematically missing in a large data set (van Buuren, 2018).
Show code
mice_model <- mice(PISA_Subsetted, method='pmm', seed = 123)
iter imp variable
1 1 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
1 2 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
1 3 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
1 4 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
1 5 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
2 1 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
2 2 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
2 3 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
2 4 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
2 5 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3 1 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3 2 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3 3 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3 4 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
3 5 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
4 1 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
4 2 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
4 3 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
4 4 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
4 5 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
5 1 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
5 2 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
5 3 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
5 4 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY
5 5 REPEAT FEMALE ESCS DAYSKIP CLASSSKIP LATE BEINGBULLIED DISCLIMA ADAPTIVITY vars n mean sd median trimmed mad min max
REPEAT* 1 20727 1.06 0.23 1.00 1.00 0.00 1.00 2.00
FEMALE* 2 22651 1.50 0.50 2.00 1.50 0.00 1.00 2.00
ESCS 3 21490 0.38 0.83 0.48 0.41 0.87 -6.69 4.04
DAYSKIP* 4 19312 1.31 0.66 1.00 1.16 0.00 1.00 4.00
CLASSSKIP* 5 19323 1.44 0.76 1.00 1.27 0.00 1.00 4.00
LATE* 6 19337 1.82 0.98 2.00 1.65 1.48 1.00 4.00
BEINGBULLIED 7 18820 0.17 1.06 0.15 0.03 1.38 -0.78 3.86
DISCLIMA 8 21353 -0.12 1.07 -0.04 -0.11 1.00 -2.71 2.03
ADAPTIVITY 9 20456 0.18 1.08 0.19 0.20 0.98 -2.27 2.01
range skew kurtosis se
REPEAT* 1.00 3.88 13.05 0.00
FEMALE* 1.00 0.00 -2.00 0.00
ESCS 10.72 -0.47 0.85 0.01
DAYSKIP* 3.00 2.41 5.80 0.00
CLASSSKIP* 3.00 1.84 2.87 0.01
LATE* 3.00 1.01 -0.06 0.01
BEINGBULLIED 4.64 0.88 0.31 0.01
DISCLIMA 4.75 -0.13 0.16 0.01
ADAPTIVITY 4.27 -0.18 -0.19 0.01Show code
psych::describe(PISA_miceimp)
vars n mean sd median trimmed mad min max
REPEAT* 1 22653 1.06 0.23 1.00 1.00 0.00 1.00 2.00
FEMALE* 2 22653 1.50 0.50 2.00 1.50 0.00 1.00 2.00
ESCS 3 22653 0.38 0.83 0.48 0.41 0.86 -6.69 4.04
DAYSKIP* 4 22653 1.32 0.67 1.00 1.17 0.00 1.00 4.00
CLASSSKIP* 5 22653 1.45 0.76 1.00 1.27 0.00 1.00 4.00
LATE* 6 22653 1.83 0.98 2.00 1.66 1.48 1.00 4.00
BEINGBULLIED 7 22653 0.18 1.07 0.15 0.04 1.38 -0.78 3.86
DISCLIMA 8 22653 -0.12 1.07 -0.04 -0.11 1.00 -2.71 2.03
ADAPTIVITY 9 22653 0.17 1.09 0.19 0.20 0.98 -2.27 2.01
range skew kurtosis se
REPEAT* 1.00 3.84 12.74 0.00
FEMALE* 1.00 0.00 -2.00 0.00
ESCS 10.72 -0.47 0.84 0.01
DAYSKIP* 3.00 2.38 5.62 0.00
CLASSSKIP* 3.00 1.83 2.81 0.01
LATE* 3.00 1.00 -0.09 0.01
BEINGBULLIED 4.64 0.88 0.35 0.01
DISCLIMA 4.75 -0.13 0.15 0.01
ADAPTIVITY 4.27 -0.17 -0.20 0.01The message above shows that the algorithm went over the data set 5 times per iteration, with the total of 25 times in 5 iterations (5 x 5). In other words, the machine imputed the missing over and over again until the change becomes minimal to give us the most stable replacement value as possible.
There is no substantial difference in descriptive statistics of the pre-imputed and post imputed data set. Given that we gained 10% of our data back, it is a win for us.
Concluding Remark
Data cleaning is a challenging, but necessary, process in data work. That is why it is important for us to know how to identify and deal with missing data appropriately before proceeding further into developing a statistical model and drawing conclusions from it. With a solid data preparation, combining with a thorough literature review, it is likely that we can draw meaningful conclusions from the data to inform our future decisions. The opposite is also true as well for poorly processed data sets. We wouldn’t want to waste our time and resources to know that the conclusion we draw is not well-supported.
A bit of controversial topic here. Non-methodologists might have some concerns that we cannot just make up the obtained scores. Like, what if the participants did not answer that question for a reason? How can we be sure that the number we generated will represent characteristics of the targeted population? The million-dollar question is, would you still do this, knowing that the number you generated might have some degree of error? Are you willing to trade authenticity of the data for the data point that might improve your statistical models? It is your task as a researcher and an informed individual to justify your choice in this matter, as well as other choices that you made in your endeavor.
Anyway, thank you so much for your read as always! Happy Holiday, everyone! I hope you have an awesome break! :)